When you’re setting up a functional genomics study, picking the right genetic manipulation tool—whether CRISPR or RNAi—can be the critical step that determines whether your project delivers breakthrough insights or stalls in confusion. Both tools have transformed biology, but understanding their real-world differences is essential to avoiding costly setbacks in your experiments.
Here’s a practical guide on choosing the right method based on your project goals, experimental constraints, and budget considerations.
Why Your Choice Matters: Real-World Implications
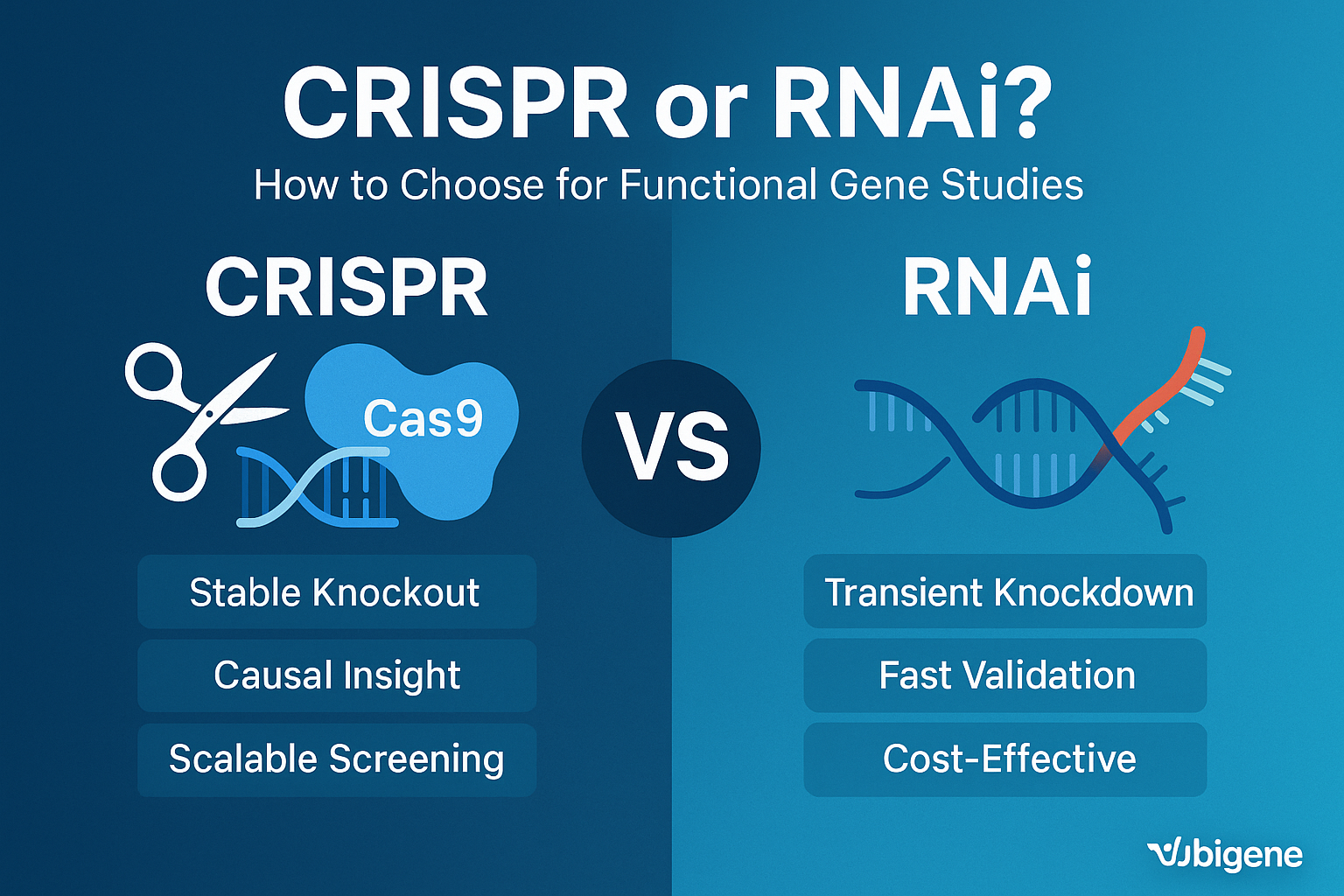
Imagine your team is working urgently to identify the genetic basis of chemotherapy resistance in lung cancer. You need a method that clearly distinguishes causal genes from bystanders. While both CRISPR and RNAi can silence genes, the mechanisms behind them differ dramatically:
- RNAi offers temporary, partial gene silencing by targeting mRNA. It’s quick and economical but known for variable knockdown efficiency.
- CRISPR gene editing directly modifies the genome, producing clear-cut gene knockouts (KO) or gene expression modulation (CRISPRi/a), resulting in precise and reproducible results.
Your choice here shapes your project’s efficiency, accuracy, and downstream validation burden.
CRISPR Gene Editing Services vs. RNAi: Functional Genomics in Practice
Let’s quickly review the strengths and limitations of each method:
| Feature | RNAi | CRISPR |
|---|---|---|
| Mechanism | mRNA degradation | DNA-level knockout/editing |
| Knockdown | Partial, transient | Permanent or controllable |
| Off-target effects | High (difficult to control) | Lower (improved gRNA design) |
| Scalability | Ideal for small screens | Highly scalable (e.g., genome-wide) |
| Validation needed | Extensive | Focused and less intensive |
| Cost | Lower initial costs | Higher upfront, better long-term ROI |
For researchers looking to explore gene editing services that support genome-wide screening,
click here to discover Ubigene’s full CRISPR gene editing capabilities.

Matching the Right Application to Your CRISPR Screen or RNAi Strategy
- Genome-wide Discovery Screening
Recommended: CRISPR screen
If your goal is broad discovery—finding novel genetic regulators at scale—CRISPR-KO or CRISPRi libraries are the gold standard.
Case study: Researchers at MD Anderson Cancer Center used genome-wide CRISPR screens to uncover critical genes responsible for resistance to immunotherapy, with over 80% validation success (Nature Medicine, 2022).
For a deeper look at how a CRISPR screen is designed and executed in real-world research environments,
click here to explore Ubigene’s genome-wide CRISPR screening platform.
Difficult-to-Transduce Cell Types
Recommended: RNAi initially, then CRISPR validation
Primary immune cells or neurons, which are difficult to infect using viral vectors, benefit from initial RNAi screens (via electroporation or lipid-mediated delivery). CRISPR-based validation can then confirm RNAi hits with lentiviral systems designed for challenging cell models.
Quick, Economical Pathway Modulation
Recommended: RNAi (siRNA or shRNA)
RNAi is best suited for rapid pathway exploration or initial hypothesis testing when budget and timelines are tight. It provides enough evidence to warrant deeper follow-up experiments.

How to Reduce Off-Target Effects: Practical Tips
Off-target effects remain a significant challenge, especially in RNAi experiments, potentially obscuring real findings or introducing false positives.
RNAi Tips:
- Validate hits using multiple siRNAs or shRNAs per gene.
- Conduct rescue experiments by reintroducing an siRNA-resistant gene variant.
CRISPR Tips:
- Use AI-enhanced gRNA design algorithms to minimize off-target cuts.
- Run GUIDE-seq or similar off-target profiling before scaling up.
Real-world example: A 2023 Science Advances paper showed optimized gRNA design reduced CRISPR off-targets by over 60%, improving screen quality significantly.
The Hidden Costs: Time, Validation, and Budget
Initial RNAi costs are low, making it attractive for small labs. However, extensive validation erodes cost advantages—false positives, phenotype noise, and replication issues pile up.
CRISPR, while more expensive upfront, often reduces validation time and delivers clearer results—crucial for translational research.
Timeline Reality:
- RNAi: Initial results in 1–2 weeks, but follow-up may stretch for months.
- CRISPR: 4–8 weeks from screen to hits, but streamlined validation.
FAQ: Real Questions from the Lab Bench
Q1: Can I start with RNAi and validate hits with CRISPR?
A: Absolutely. Many labs use RNAi for early-stage discovery and follow up with CRISPR for definitive confirmation.
Q2: How many replicates do I need?
A: At least three biological replicates for either method ensures statistical reliability and reduces false positives.
Q3: Is CRISPR always better for validation?
A: Not always. For essential genes, partial knockdown via RNAi or CRISPRi can offer more biologically relevant results than full knockouts.
Validation: The Crucial Step You Can’t Afford to Skip
Validation is non-negotiable, especially with RNAi, which often demands:
- Multiple siRNAs per gene
- Rescue experiments
- Confirmation with orthogonal tools (e.g., CRISPRi, CRISPRa, inhibitors)
By contrast, CRISPR’s genotype-to-phenotype clarity reduces validation complexity—but still benefits from orthogonal confirmation.
Case insight: Nature Biotechnology (2023) reported ImmunoBioTech needed twice as much validation for RNAi-derived hits compared to CRISPR hits.
Emerging Use Cases: When the Lines Blur
In newer research areas such as drug repurposing, epigenetic regulation, and non-coding RNA function, the choice between RNAi and CRISPR isn’t always binary—many teams now combine them for iterative insight.
Drug Repurposing Screens
Repurposing existing FDA-approved drugs for new indications often requires rapid validation of compound-gene interactions. RNAi offers a quick first-pass approach to identify pathway involvement, while CRISPR screens confirm whether knocking out a target replicates or blocks the drug effect.
Strategy Tip: Start with RNAi for speed, then layer in CRISPR to de-risk repurposing candidates.
Epigenetic and Non-coding RNA Studies
CRISPRi (interference) and CRISPRa (activation) have opened new doors in studying regulatory elements, enhancers, and lncRNAs. Unlike RNAi, which struggles with chromatin-modifying targets, CRISPRi/a can modulate gene expression without altering the sequence—ideal for studying epigenetic silencing, promoter accessibility, or transcriptional rewiring.
Tool Pairing Tip: Combine CRISPRi with chromatin immunoprecipitation (ChIP) assays or RNA-seq to dissect complex regulatory networks.
Functional Redundancy Testing
Some gene families exhibit redundancy—RNAi often fails to knock down all isoforms simultaneously. CRISPR multiplexing (targeting multiple genes at once) enables direct knockout of entire gene clusters, revealing phenotypes otherwise masked.
Final Decision Checklist: Choose with Confidence
| Ask Yourself… | Best Method |
|---|---|
| Need full knockout? | CRISPR-KO |
| Working with essential genes? | CRISPRi or RNAi |
| Budget-limited and need fast results? | RNAi |
| Prioritize reproducibility and validation ease? | CRISPR |
| Hard-to-transduce cells? | RNAi first, then CRISPR |
| Translational pipeline downstream? | CRISPR |
Conclusion: Strategy Over Tools
Ultimately, this isn’t CRISPR vs. RNAi—it’s about aligning your tools with your goals. Smart experimental design means:
- Start with a clear biological question
- Choose the most appropriate platform
- Plan for layered validation
- Use both tools strategically when needed
When properly applied, RNAi offers speed, and CRISPR delivers precision. Together, they form a powerful toolkit for modern functional genomics.
Want to explore genome-scale screening with CRISPR?
Start with Ubigene’s CRISPR screen platform—it’s purpose-built for discovery at scale.